Project #1: Copper Chaperones
Background:
Copper ions serve as essential
co-factors for metalloproteins (e.g. for those involved in energy generation
and iron uptake and distribution), but at the same time are potentially toxic
to living cells due to their ability to bind with high affinity to partially
folded proteins, their high redox activity, and their propensity to catalyze
the auto-oxidation of lipids, proteins, and nucleic acids. Copper chaperone proteins have consequently
evolved as part of a complex network for the intracellular trafficking of
copper and help to control the amount of free intracellular copper. These proteins not only aid in establishing a
homeostatic balance of this essential but toxic metal, but also help to ensure
proper delivery of the ions to the metalloprotein(s) where catalysis ultimately
takes place. Maintenance of
intracellular copper ion concentrations by these chaperone proteins is not
presently well understood and constitutes an important problem in bioinorganic
chemistry.
Systems of Interest:
The copper chaperone proteins Atx1
(in yeast) and Hah1 (in humans) are small soluble copper-binding proteins which
deliver CuI to the copper-transporting P-type ATPase molecules Ccc2 in
yeast and Wilson (WND) or Menkes (MNK) disease proteins in humans, which in
turn translocate CuI into the lumen of the Golgi in cells for
loading into the multicopper oxidase Fet3.
A largely conserved mechanism for Cu acquisition and distribution from yeast
to humans makes these systems ideal for a dual study. Interactions between copper chaperones and
ATPases are also of particular interest as mutations in genes coding ATPases
have been implicated in inherited disorders of copper metabolism and neurodegenerative
disorders. Mutations in MNK lead to a
copper deficiency disease (Menkes’ disease) and in WND to a copper toxicity
condition (Wilson’s disease).
While no biomimetic models have been
constructed for these proteins, the structures of apo- and CuI-loaded
Atx1 have been solved by NMR spectroscopy.
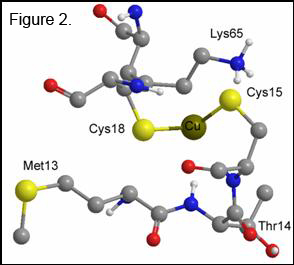
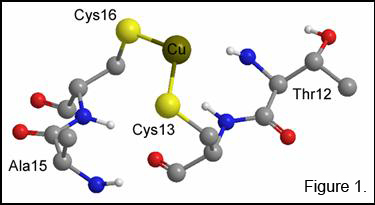
The CuI center is coordinated by two cysteine residues (Cys15
and Cys18) and the S–Cu–S bond angle suggests the metal should be 3-coordinate
(Figure 1). Uncertainty exists
concerning the identity of the third ligand, which could be a conserved
methionine residue, an exogenous thiol group (most likely), or an oxygen or
nitrogen from a spatially close amino acid residue. EXAFS data predicts that it is a third S atom
which completes the CuI coordination sphere. The metal binding site is exposed at the
protein surface, suiting its purpose of delivery rather than catalysis or
electron transfer. The structure of the
first domain of Ccc2 has likewise been solved by NMR spectroscopy and shows the
ATPase to have the same CXXC Cu-binding motif as the chaperone protein (Figure
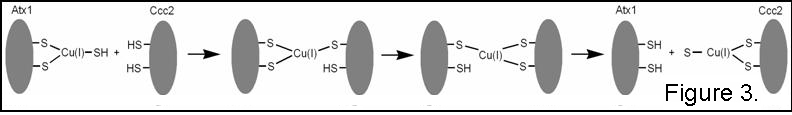
2). Cu transfer from Atx1 to Ccc2 has
been shown to have Keq = 1.5, implying transfer is not based on a
higher Cu affinity in the target domain, and kex > 103
s-1. This has led to the
proposal of a mechanism where metal ion exchange occurs through the formation
of two- and three-coordinate intermediates involving cysteine residues from
both proteins (Figure 3).
Human Hah1 protein structures reveal
that, unlike in Atx1, the CuI center has a distorted tetrahedral
coordination geometry with four ligating cysteines, which has not been reported
before for biological systems. HgII-
and CdII-bound forms of Hah1 were also determined and seen to have
different binding modes, which is suspected to be responsible for their
differing rates of metal transfer to ATPases.
The structure for the fourth metal-binding domain from the Menkes
copper-transporting ATPase has been solved as well.
Research Goals and Projects:
![]() Computations
will first be used to address how different metals (CuI, HgII,
CdII) bind to the Cu chaperone Atx1 and the ATPase Ccc2. Data from the CopZ bacterial copper chaperone
suggest that metal binding affinity should follow the order CuI >
HgII > CdII. In
studying these metal-bound complexes, the effect of differing the coordination
number and ligands will be explored.
Results will lead to understanding what type of coordination environment
best stabilizes CuI, determination of the third ligand in Atx1, and
establish a basis for differing metal binding affinities.
Computations
will first be used to address how different metals (CuI, HgII,
CdII) bind to the Cu chaperone Atx1 and the ATPase Ccc2. Data from the CopZ bacterial copper chaperone
suggest that metal binding affinity should follow the order CuI >
HgII > CdII. In
studying these metal-bound complexes, the effect of differing the coordination
number and ligands will be explored.
Results will lead to understanding what type of coordination environment
best stabilizes CuI, determination of the third ligand in Atx1, and
establish a basis for differing metal binding affinities.
![]() At this
point, the mechanism of metal transfer from Cu chaperone to ATPase can be
investigated. The presently proposed
mechanism and possible alternative mechanisms can be characterized, with
elucidation of energetics and structures of intermediates and transition
states.
At this
point, the mechanism of metal transfer from Cu chaperone to ATPase can be
investigated. The presently proposed
mechanism and possible alternative mechanisms can be characterized, with
elucidation of energetics and structures of intermediates and transition
states.
![]() This work
can be followed by analogous calculations with Hah1 and the Menkes ATPase. Both the metal binding and metal transfer
studies will initially be carried out on quantum (QM) model structures
extracted from the available protein structures. Results obtained thereby can also be
contrasted with experimental data from the full protein systems to gauge the effects
of the protein environment on the chemistry.
This work
can be followed by analogous calculations with Hah1 and the Menkes ATPase. Both the metal binding and metal transfer
studies will initially be carried out on quantum (QM) model structures
extracted from the available protein structures. Results obtained thereby can also be
contrasted with experimental data from the full protein systems to gauge the effects
of the protein environment on the chemistry.
![]() Subsequently, QM/MM calculations can be
performed to directly access chemistry in the proteins. Residues can be mutated in computations and
their effects on CuI-binding and transport properties measured.
Subsequently, QM/MM calculations can be
performed to directly access chemistry in the proteins. Residues can be mutated in computations and
their effects on CuI-binding and transport properties measured.
Reading
(1) Luk, E.; Jensen,
L. T.; Culotta, V. C. J. Biol. Inorg.
Chem. 2003, 8, 803-809.
(2) Arnesano, F.;
Banci, L.; Bertini,
(3) Harrison, M. D.;
Jones, C. E.; Dameron, C. T. J. Biol.
Inorg. Chem. 1999, 4, 145-153.
(4) Huffman, D. L.;
O'Halloran, T. V. Annu. Rev. Biochem.
2001, 70, 677-701.
(5) Lu, Z. H.; Solioz,
M. Adv. Protein Chem. 2002, 60, 93-121.
(6) Elam, J. S.;
Thomas, S. T.; Holloway, S. P.; Taylor, A. B.; Hart, P. J. Adv. Protein Chem. 2002,
60, 151-219.