Squishy Physics Research
Our research primarily focuses on the physical characterization of soft condensed matter systems, such as biological polymer networks, cells, tissues, and biofilms. The non‑equilibrium nature and rich phase space of active soft condensed matter make for complex physical systems with many layers of emergent behavior. We use both computational and experimental approaches to dissect the underlying physical principles of complex soft matter systems. Below is an overview of some of our past and current research projects.
Current Projects
Lipid/polymer model systems
Living cells are dynamic systems operating far from thermal equilibrium. Cells dynamically change shape and alter their physical and chemical properties in response to their local microenvironment. These shape changes involve both alterations of the cell's cytoskeleton, the internal network of biopolymers which grant the cell its mechanical properties, and the lipid cell membrane surrounding the cellular interior.
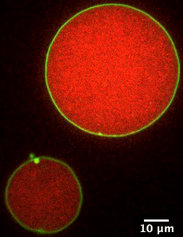
We design lipid/polymer model systems to study the physical interplay between encapsulated filaments and the encapsulating lipid membrane, and are developing quantitative methods to analyze the energetics of their assembly from confocal fluorescence imaging. We also utilize bulk rheology to characterize the mechanics of the polymer interior. This "bottom-up" approach uses either reconstituted filamentous proteins (actin; shown in red in the image) or non-biological polymers encapsulated in giant unilamellar vesicles (DOPC; shown in green in the image), and allows us to probe the underlying physics and interactions between the components. This is an ongoing collaboration with the Parikh lab at UC Davis.
Relevant reading: Su et al., 2023.
Rheological characterization of plastics degradation
Over 1/5 of waste from the roughly 300 million tons of plastic materials, produced globally each year, ends up in our oceans, rivers, and creeks. Varied environmental conditions such as exposure to UV light and diverse salinities, degrade these plastics into microplastics which then become integrated into the ecosystems and can enter the food chain.
This work focuses on using rheological techniques to mechanically characterize plastic samples as they are subjected to degradation factors such as UV light and high salinity, and compare them to bio-compatible plastic alternatives. We use our lab's high sensitivity force-extension rig to characterize the viscoelasticity of plastics and plastic alternatives to better understand how environmental factors affect their mechanical degradation.
Fruit fly smooth muscle mechanics and autism spectrum disorder

Our bodies are resident to vast populations of microbial species, and it has become increasingly clear that the symbiotic relationships we share with our resident microbes are critical for human development and adult homeostasis. For example, emergent data indicates that manipulating the gut microbiome can impact neural development and function. Gastrointestinal problems in humans are comorbid with a number of neuropsy chiatric disorders, including autism spectrum disorder (ASD), and recent clinical trials suggest manipulation of the gut microbiome can affect ASD symptoms. We characterize the mechanics of intestinal smooth muscle tissue dissected from Drosophila (a genus of flies) carrying ASD-related gene mutations. The smooth muscle tissue (comparable in thickness to human hair, but much softer) is suspended between two clips and extended using our lab's high-sensitivity force-extension rig. The aim is to investigate the reciprocal relationship between neural development and intestinal health from a biomechanical perspective by combining biological genetic techniques and genetic characterizations with rheological measurements.
Relevant reading: Diamond et al., 2011; Clemente et al., 2012; Swank, 2012; Rosenfeld, 2015; Sampson & Mazmanian, 2015; Cryan et al., 2019.
Past Projects
Protein self-assembly
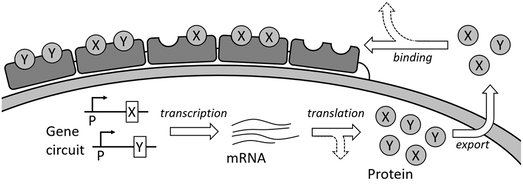 Protein complexes consisting of multiple subunits are assembled both inside and outside cell through tight coordination of gene expression, protein transport, and binding processes. Since the cellular protein expression machinery involves a relatively small number of ensemble proteins, the system is ill described by steady state thermodynamics. We therefore use a combination of computational Monte Carlo simulations and theoretical modeling to describe the protein assembly structures. This project focuses on how the genetic circuits and dynamic rates affect the final non-steady state protein assembly structure, and highlights similarities in dynamic arrest between other non-steady state assemblies, such as reconstituted biopolymer networks. The work is a collaboration with the Tan lab at UC Davis.
Protein complexes consisting of multiple subunits are assembled both inside and outside cell through tight coordination of gene expression, protein transport, and binding processes. Since the cellular protein expression machinery involves a relatively small number of ensemble proteins, the system is ill described by steady state thermodynamics. We therefore use a combination of computational Monte Carlo simulations and theoretical modeling to describe the protein assembly structures. This project focuses on how the genetic circuits and dynamic rates affect the final non-steady state protein assembly structure, and highlights similarities in dynamic arrest between other non-steady state assemblies, such as reconstituted biopolymer networks. The work is a collaboration with the Tan lab at UC Davis.
Relevant reading: Jensen et al., 2020.
F-actin tropomyosin interactions
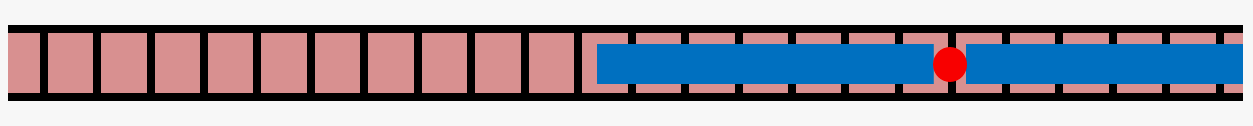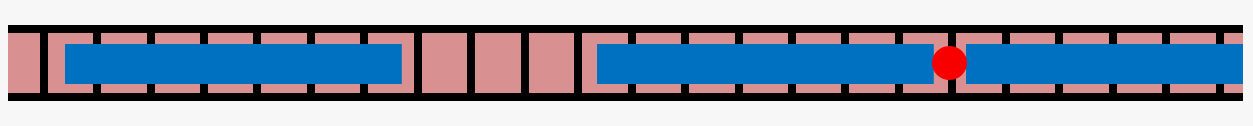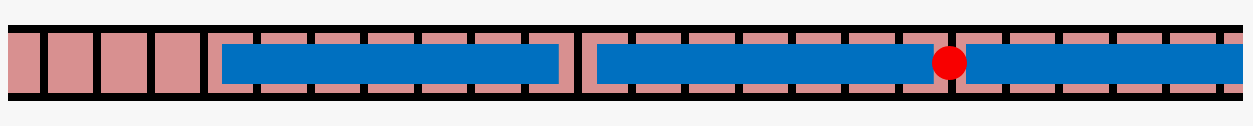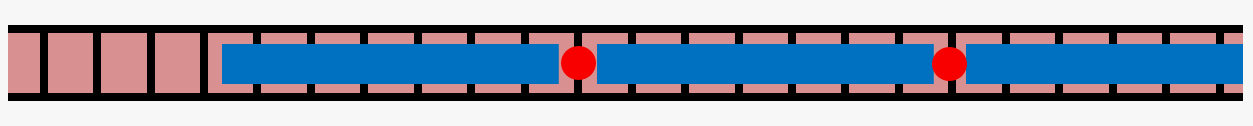 Actin is a cytoskeletal protein capable of polymerizing into semiflexible filaments, which play a crucial role in determining cell mechanics. Filamentous actin (shown in red in the cartoon below) is regulated by a range of actin-binding proteins, which regulate its chemical and physical properties.
Actin is a cytoskeletal protein capable of polymerizing into semiflexible filaments, which play a crucial role in determining cell mechanics. Filamentous actin (shown in red in the cartoon below) is regulated by a range of actin-binding proteins, which regulate its chemical and physical properties.
One such protein, tropomyosin (shown in blue), binds to polymerized actin in a 1:7 ratio, with one tropomyosin unit occupying 7 actin monomers, to form tropomyosin chains on filamentous actin. This leaves open the possibility of "gaps" in decoration of fewer than 7 actin monomers between segments of tropomyosin. We are exploring lateral diffusion of tropomyosin as a possible mechanism for resolving these gaps using theoretical modeling and computational Monte Carlo simulations.
Relevant reading: Fischer et al., 2016; Janco et al., 2020; Jensen et al., 2020.
Myosin motility assays
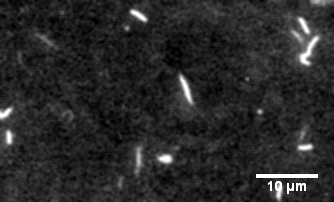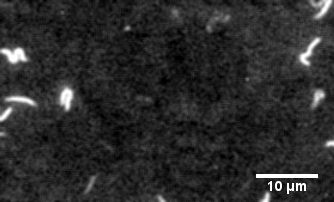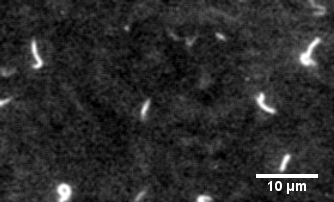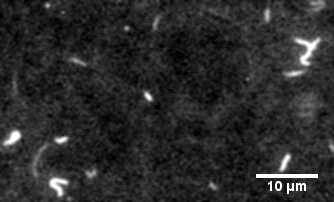
The protein primarily responsible for force generation in your muscles is myosin II, a molecular motor which tugs on actin filaments to generate force and contraction. The myosin in vitro motility assay is used to characterize the molecular motor myosin and its interaction with filamentous actin. In the assay, a bed of myosin motors are laid on a glass surface, and fluorescently labeled F-actin filaments are imaged as the myosin motors propel them across the surface.
The assay allows for the physical characterization of the calcium regulation, force production, and power output of myosin and related disease mutants, as well as actin-binding proteins as they interact with the F-actin filaments being strained by the myosin motors.
Relevant reading: Warshaw, 1996; Greenberg et al., 2010.
Biopolymer and cell rheology
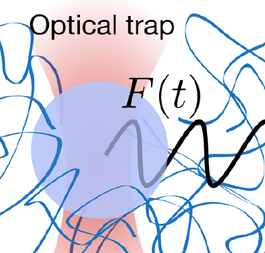
Mechanics and mechanical processes play a crucial role in biology. We use both microrheology and bulk rheology to study biological systems, such as living cells or microbial aggregates, and reconstituted model systems, such as polymer networks. Bulk rheology probes the mechanics of a macroscopic sample by applying a shear force and measuring the corresponding strain. To probe local mechanics, a focused laser beam can be used to manipulate transparent microscopic particles: the dielectric material refracts the light, and the focused laser beam acts as a soft spring, exerting forces in the pico-Newton range and pulling dielectric material towards the laser focus. By moving the laser beam and tracking the corresponding bead motion, these "laser tweezers" can be used to probe the mechanics of the surrounding environment.
Relevant reading (microrheology): Guo et al., 2013; Guo et al., 2014; Guo et al., 2017.
Relevant reading (bulk rheology): Jensen, Morris, et al., 2014; Jensen, Morris, et al., 2014.